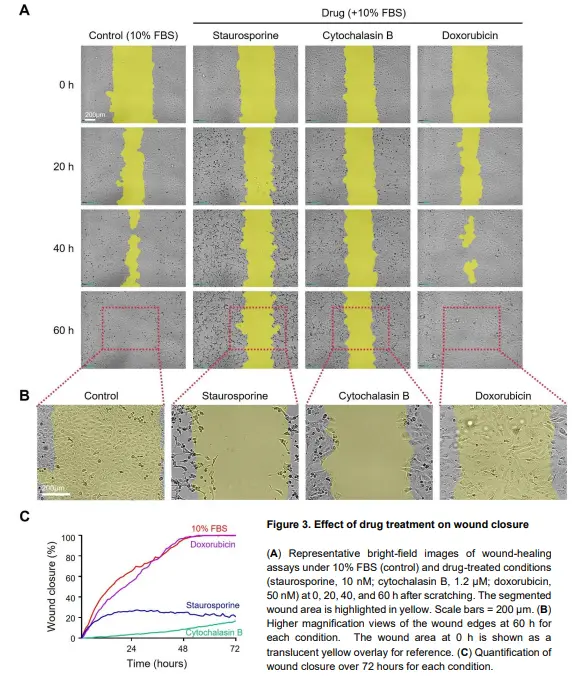
In addition to serum conditions, wound closure can be influenced by cell migration, proliferation, and cell
death. To examine how perturbing these processes alters closure kinetics, additional wound-healing assays
were performed with staurosporine, cytochalasin B, and doxorubicin at sublethal concentrations.
In representative images (Figure 3A), the 10% FBS control condition showed robust wound closure. In
contrast, staurosporine, a potent apoptosis inducer2
, markedly reduced wound-healing activity. Cells
displayed progressive morphological changes, including shrinkage, consistent with apoptosis (Figure 3A-B).
Cell death became evident around 20 hours, causing the wound closure curve to plateau, with little to no
further closure observed thereafter (Figure 3C).
Cytochalasin B also markedly reduced wound closure. Because cytochalasin B disrupts actin polymerization
and impairs the cytoskeletal dynamics required for motility3
, this effect is consistent with reduced migratory
capacity. No evident cell death was observed, but proliferation appeared suppressed (Figure 3B), limiting
advancement of the leading edge and slowing closure (Figure 3C).
Interestingly, doxorubicin produced a wound closure rate similar to the control. As a chemotherapeutic agent
that induces DNA damage4
, doxorubicin did not cause pronounced cell death under the conditions tested but
appeared to suppress cell proliferation. Despite this growth inhibition, wound closure remained comparable
to the control because cells showed extensive spreading while maintaining migratory activity. This suggests
that suppressing proliferation alone may not be sufficient to reduce wound closure and that effective inhibition
requires direct impairment of migration. Notably, under mild cytotoxic stress, doxorubicin may promote a
compensatory response in which cells increase their footprint and continue collective migration despite
reduced growth5
. These results demonstrate that wound closure curves vary markedly depending on the drug
applied and can be categorized into distinct patterns (e.g., steep upward, gradual upward, or flat) based on
their trends.
Taken together, these findings highlight the value of live-cell imaging for interpreting wound closure dynamics,
as migration- and proliferation-associated behaviors can contribute differently depending on the condition or
treatment. Distinguishing the relative contributions of these processes enables more accurate assessment
of wound-healing activity and provides clearer insight into how external stimuli modulate collective cell
behavior.
In addition to serum conditions, wound closure can be influenced by cell migration, proliferation, and cell
death. To examine how perturbing these processes alters closure kinetics, additional wound-healing assays
were performed with staurosporine, cytochalasin B, and doxorubicin at sublethal concentrations.
In representative images (Figure 3A), the 10% FBS control condition showed robust wound closure. In
contrast, staurosporine, a potent apoptosis inducer2
, markedly reduced wound-healing activity. Cells
displayed progressive morphological changes, including shrinkage, consistent with apoptosis (Figure 3A-B).
Cell death became evident around 20 hours, causing the wound closure curve to plateau, with little to no
further closure observed thereafter (Figure 3C).
Cytochalasin B also markedly reduced wound closure. Because cytochalasin B disrupts actin polymerization
and impairs the cytoskeletal dynamics required for motility3
, this effect is consistent with reduced migratory
capacity. No evident cell death was observed, but proliferation appeared suppressed (Figure 3B), limiting
advancement of the leading edge and slowing closure (Figure 3C).
Interestingly, doxorubicin produced a wound closure rate similar to the control. As a chemotherapeutic agent
that induces DNA damage4
, doxorubicin did not cause pronounced cell death under the conditions tested but
appeared to suppress cell proliferation. Despite this growth inhibition, wound closure remained comparable
to the control because cells showed extensive spreading while maintaining migratory activity. This suggests
that suppressing proliferation alone may not be sufficient to reduce wound closure and that effective inhibition
requires direct impairment of migration. Notably, under mild cytotoxic stress, doxorubicin may promote a
compensatory response in which cells increase their footprint and continue collective migration despite
reduced growth5
. These results demonstrate that wound closure curves vary markedly depending on the drug
applied and can be categorized into distinct patterns (e.g., steep upward, gradual upward, or flat) based on
their trends.
Taken together, these findings highlight the value of live-cell imaging for interpreting wound closure dynamics,
as migration- and proliferation-associated behaviors can contribute differently depending on the condition or
treatment. Distinguishing the relative contributions of these processes enables more accurate assessment
of wound-healing activity and provides clearer insight into how external stimuli modulate collective cell
behavior.
Conclusion
This application note highlights the effectiveness of the Celloger® Pro system for wound-healing assays.
Using NIH-3T3 cells, the system clearly detected differences in wound closure rates across FBS
concentrations and enabled classification of distinct drug-induced closure patterns. By automating both
image acquisition and wound area quantification, the system enables high-resolution, time-dependent
monitoring of wound closure with minimal manual intervention.
Notably, the advanced wound detection algorithm provides accurate and consistent segmentation of the
wound region, even in low-contrast or partially closed gaps. This improvement enhances the reliability of
quantitative analysis and reduces user-dependent variability. Overall, the ability to clearly distinguish wound
closure responses under different conditions underscores the value of Celloger® Pro as a powerful all-in-one
solution for regenerative medicine research and cell motility studies.
Conclusion
This application note highlights the effectiveness of the Celloger® Pro system for wound-healing assays.
Using NIH-3T3 cells, the system clearly detected differences in wound closure rates across FBS
concentrations and enabled classification of distinct drug-induced closure patterns. By automating both
image acquisition and wound area quantification, the system enables high-resolution, time-dependent
monitoring of wound closure with minimal manual intervention.
Notably, the advanced wound detection algorithm provides accurate and consistent segmentation of the
wound region, even in low-contrast or partially closed gaps. This improvement enhances the reliability of
quantitative analysis and reduces user-dependent variability. Overall, the ability to clearly distinguish wound
closure responses under different conditions underscores the value of Celloger® Pro as a powerful all-in-one
solution for regenerative medicine research and cell motility studies.